Myriopteris aurea (Poir.) Grusz & Windham
I. Plant Systematics
The evolution of apomixis in ferns
Apomixis is asexual reproduction through seed, spore, or egg, without the fusion of gametes [“apo” = without, “mixis” = mixing/sex]. In plants, this definition encompasses vegetative asexual reproduction and asexual reproduction through the alternation of generations (including apospory, apogamy, and parthenogenesis). The Grusz lab the evolution (and evolutionary implications) of apogamous reproduction, in particular. In ferns, this involves two major modifications to the canonical, sexual life cycle: the production of unreduced diploid spores via meiosis (diplospory), and the apogamous development of a new sporophyte from somatic gametophyte tissue, without the fusion of sperm and egg (apogamy). Sporogenesis takes place within the fern sporangium, where spores develop via meiosis. In sexual lineages, these meiotic products (spores) are normally haploid, with half the parental number of chromosomes. Instead, apomictic ferns use one of two alternative spore-generating pathways to yield chromosomally unreduced, i.e. diploid, spores: premeiotic endomitosis (PE; Fig. B.i.) and meiotic first division restitution (MFDR; Fig. B.ii.).
> Grusz. 2016. Journal of Systematics and Evolution.
apogamous fern life cycle (left). Sporogenesis via (i.) Premeiotic endomitosis (= Döpp-Manton) and (ii) meiotic first division restitution (= braithwaite) in ferns (right).
Reticulate evolution and origins of apomixis
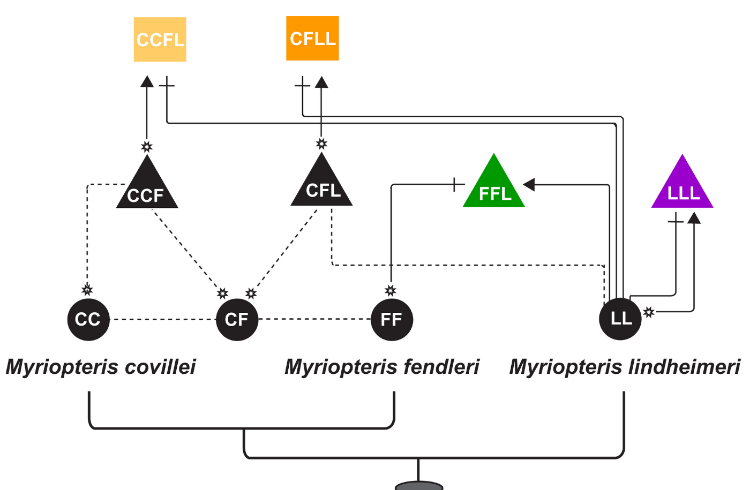
Evolutionary relationships in the CheilantHes (= Myriopteris) yavapensis species complex.
We use a variety of experimental approaches—spanning cytogenetics (chromosomes!) to phylogenetics (character evolution!)—to explore the evolutionary origins and the evolutionary consequences of asexual (apomictic) reproduction in ferns. We leverage spore studies, flow cytometry, and chromosome counts to determine,
How many full genome copies interact in each cell nucleus?
Enzyme electrophoresis, along with DNA/RNA sequencing, are used to expose,
Which divergent genomes comprise each nucleus, in what dosage? and
What (if any) are the short- and long-term evolutionary effects?
Together with morphological data, comparative studies such as these are illuminating (often reinforcing ) the evolutionary origin(s) of apomictic polyploid hybrids.
> Grusz et al. 2009. American Journal of Botany.
Exploring the evolution of apomictic lineages
chromosome number, ploidy level, and reproductive mode across Myriopteris (Pteridaceae). C = covillei clade, L = lanosa clade, A = alabamensis clade.
Morphological convergence, together with polyploidy, hybridization, and apomixis, can complicate efforts to infer phylogenetic relationships in some plant lineages, usually resulting in severe taxonomy instability. Cheilanthoid ferns (Pteridaceae), Cheilanthes Sw. in particular, are a classic example of the taxonomic instability that can result from such confounding factors (Windham et al. 2009). Phylogenetic analyses of cheilanthoid ferns have demonstrated the genus Cheilanthes to be grossly polyphyletic, with species assigned to the name dispersed over five of six major cheilanthoid clades (Windham et al. 2009). In an effort to stabilize taxonomy, we resurrected the genus, Myriopteris Fée; Grusz and Windham 2013), thereby reflecting an improved understanding of evolutionary history. Our phylogenetic analyses found strong statistical support for intrageneric species relationships, and shed light on morphological, cytological, and reproductive character evolution in the group (Grusz et al. 2014).
> Grusz et al. 2014. Systematic Botany.
> Grusz and Windham. 2013. Phytokeys.
> Windham et al. 2009. American Fern Journal.
II. Molecular Marker Development
Bulk identification of single copy nuclear gene regions (RNA-Seq)
PIPELINE FOR THE identifICATION OF single copy orthologous regions.
The sequencing of transcribed RNA can aid greatly in the simplification of complex and/or repetitive genomes. Leveraging bioinformatic tools, these data facilitate a variety of molecular studies, including whole genome assembly and low-copy molecular marker development. Using data gathered for the 1KP Project (onekp.com), this project utilized high-throughput bioinformatic pipelines to identify and extract >2000 orthologous gene regions from Illumina PE RNA-Seq data from species spanning the fern family Pteridaceae.
> Grusz et al. 2016. BMC Genomics.
Repetitive DNA: simple sequence repeats (SSRs)
polymorphic microsatellite (SSR) marker: (GCC)^3 and (GCC)^6.
Ferns are notorious for their large, repetitive genomes. We exploit this tendency to develop genetic markers that are polymorphic for DNA repeat length. With these molecular tools, we examine genetic and genotypic variation in nature to shed light on micro- and macroevolutionary processes.
> Grusz and Pryer. 2014. Applications in Plant Sciences.
Low-copy nuclear DNA markers
MAP OF NUCLEAR gapCp in ferns.
In organisms with large genomes and frequent polyploidy (genome duplication), sequence data for low copy genes can sometimes be in short supply. We use traditional and next generation sequencing approaches to develop DNA markers that are used to inform on genotypes / phenotypes, and provide insight into the evolutionary origins of polyploids.
> Schuettpelz et al. 2008. Systematic Botany.
III. Evolutionary Genomics
Mobile elements shape plastome evolution in ferns
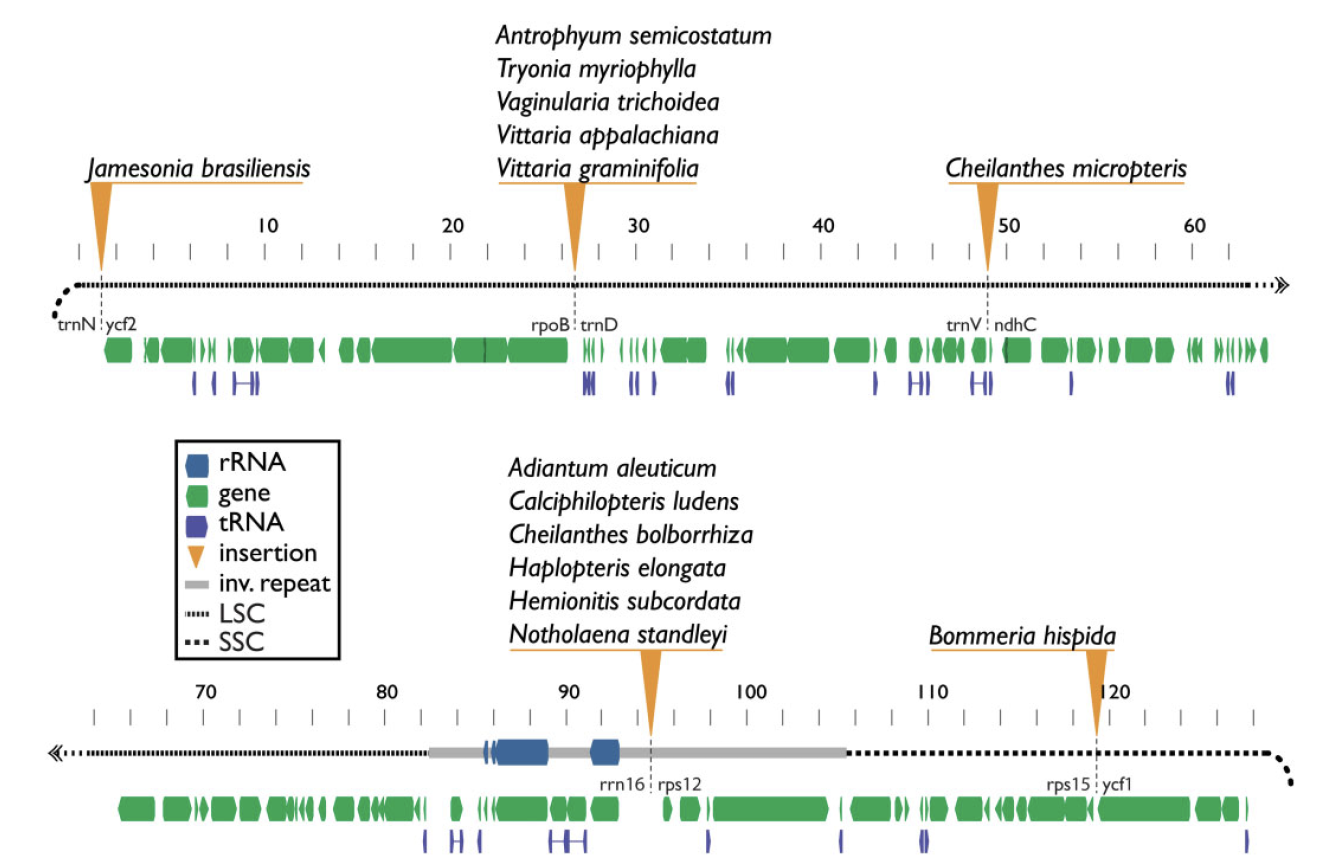
Plastid genomes display remarkable organizational stability over evolutionary time. From green algae to angiosperms, most plastid genomes are largely collinear, with only a few cases of inversion, gene loss, or, in extremely rare cases, gene addition. These plastome insertions are mostly clade-specific and are typically of nuclear or mitochondrial origin. Here, we expand on these findings and present the first family-level survey of plastome evolution in ferns, revealing a novel suite of dynamic mobile elements. Comparative plastome analyses of the Pteridaceae expose several mobile open reading frames that vary in sequence length, insertion site, and configuration among sampled taxa. Even between close relatives, the presence and location of these elements is widely variable when viewed in a phylogenetic context. We characterize these elements and refer to them collectively as Mobile Open Reading Frames in Fern Organelles (MORFFO). We further note that the presence of MORFFO is not restricted to Pteridaceae, but is found across ferns and other plant clades. MORFFO elements are regularly associated with inversions, intergenic expansions, and changes to the inverted repeats. They likewise appear to be present in mitochondrial and nuclear genomes of ferns, indicating that they can move between genomic compartments with relative ease. The origins and functions of these mobile elements are unknown, but MORFFO appears to be a major driver of structural genome evolution in the plastomes of ferns, and possibly other groups of plants.
> Robison et al. 2018. Genome Biology & Evolution
Insertion locations of MORFFO elements in members of the Pteridaceae.
Extreme evolutionary rate variation in ferns
Rapid molecular evolutionary rates in the morphologically extreme vittarioid ferns.
In the fern tree of life, some lineages evolve at mostly consistent rates (relative to one another), but others have vastly different evolutionary rates compared to closely related groups (most notably, the filmy ferns, tree ferns, and vittarioid ferns). Using transcriptome / gene expression (RNA) data, we study evolutionary rate discrepancies among the genomes of taxa spanning the Pteridaceae — a cosmopolitan fern family with ca. 50 genera and > 1000 species (ca. 10% of documented fern diversity). In collaboration with E. Schuettpelz (Smithsonian Institution, NMNH) and C. Rothfels (UC Berkeley), with support from the 1KP Project (onekp.com), our goal is to uncover genomic clues driving disparate morphologies, ecologies, life histories, and rates of molecular evolution across the family. This is part of an ongoing project with K. M. Pryer (Duke University) aimed toward identifying drivers of rapid molecular and morphological divergence between Adiantum and the vittarioid ferns, and sheds light on the mechanisms influencing evolutionary rates across the family.